
Phylogenies and Visualization
A phylogenetic tree is a diagram that illustrates how species or groups have evolved from common ancestors over time. In a time-calibrated phylogenetic tree, the edges have lengths, which can be interpreted as the time taken to evolve from the ancestors to the descendants.
Phylogenies can be visualized in many distinct layouts. While these layouts have difference appearances, they all maintain identical topological relationship between nodes and edges.

Phylogeny Is Graph
Phylogenetic trees are essentially graphs. In a phylogeny, each node represents a species or a common ancestor, and each edge represents the evolutionary connection between them.
Below I provide you an interactive playground, in which you can explore the forms of a phylogeny/graph.
You can specify a tip number and click on the "Generate Tree" button below to simulate a phylogenetic tree under the Yule model.
Click on "Phlogeny Form" and "Graph Form" to switch between common layouts of phylogeny and graph. Click on the rotation buttons to display the tree in a different pose. If you like, you can also drag the nodes around.
Enter number of tips:
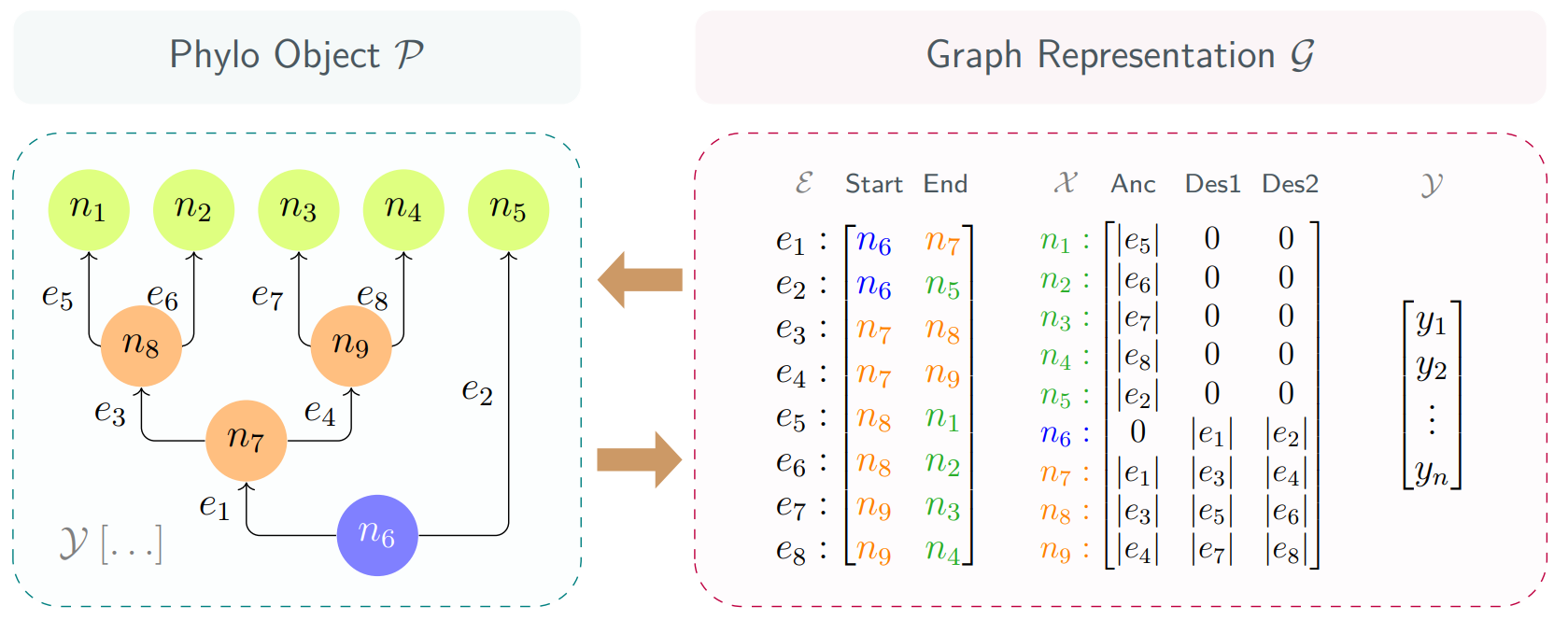
You migh have noticed two things: first, the edges are directed and represented by arrows; second, below the interactive graph there are an Adjacency List and a Node Features matrix.
In the context of phylogenies, edges are typically considered undirected, however, conventional phylogeny encoding contains only single-directional edges pointing from the ancestor nodes to their descendants. Popular deep learning frameworks recognize edges under such encoding as directed.
How exactly are phylogenies encoded conventionally? The core idea is to maintain an adjacency list. The list stores all the edges in the format [ni, nj], where ni and nj are the starting and ending nodes of the edge.
When you hover over an edge or node in the graph, its associated entries in the adjacency list will be highlighted. Similarly, hovering over a row or element in the list will highlight the corresponding node or edge in the tree. This interactive highlighting helps to easily trace connections and understand how an adjacency list sufficiently encodes a phylogeny.
You can ignore the node feature matrix for now an go back later after you read the next section.
Bridging Phylogenies and GCNs
Graph Convolutional Networks (GCNs) are a type of neural network specifically designed to operate on graph-structured data. Unlike traditional convolutional networks that work on regular grid data like images, GCNs can process data represented as nodes and edges. The core idea of GCNs is to aggregate features from a node's neighbors to capture both local graph topology and feature information. This is achieved through convolutional operations that combine and transform node features based on their connections. Visit Thomas Kipf's original blog for more information.
I proposed a protocol to directly embed the evolutionary relationships (lengths of adjacent edges) into the nodes as their features. This protocol does not require the involvement of edge features in computation.
Each row of the node feature matrix stores features of a node in the format [ei, ej, ek], where ei is the length of the edge connecting the node to its ancestor, ej and ek are the lengths of the edges connecting the node to its two descendants.
Click Here to go back to the playground and revisit the node feature matrix.